Microbial Cell Physiology and Mechanics
Cell Division & Developmental Growth Programs
Understanding the Molecular Mechanism of Cell Division
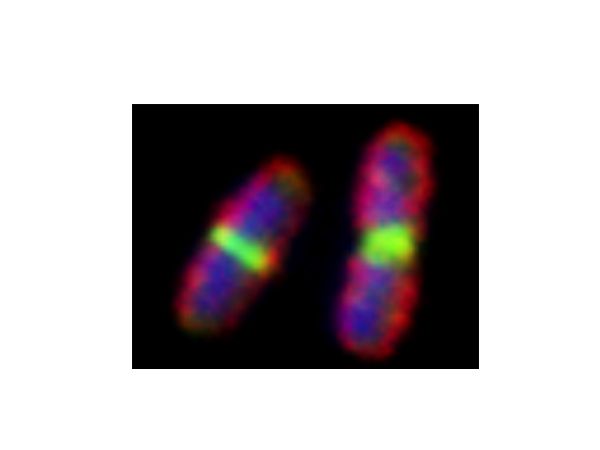
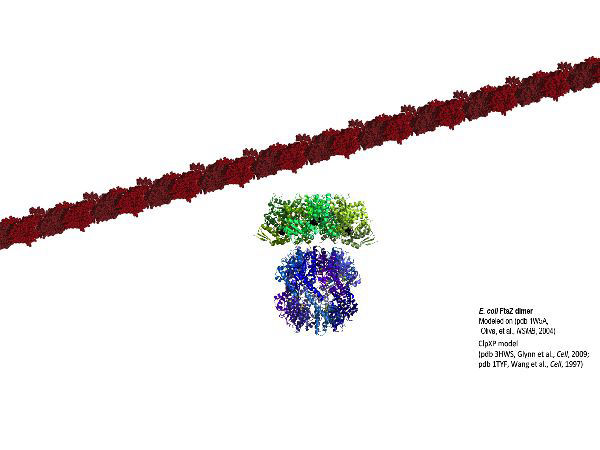
FtsZ is a highly conserved bacterial cell division protein that is a structural homolog of tubulin and polymerizes to form a dynamic protein structure called the “Z-ring” at midcell. At the Z-ring, the center of an E. coli cell "constricts" to divide into two identical progeny cells and FtsZ is essential for this process. Using in vivo and in vitro assays, we investigate proteins that recruit, stabilize and destabilize the Z-ring, as well as systems responsible for precise timing and placement of the Z-ring in vivo. The actin-like ATPase FtsA polymerizes, coassembles with FtsZ polymers, and together they form they nascent Z-ring at the site of division. We are developing methods to monitor FtsA-FtsZ complexes and determine how these complexes contribute to initation of cell wall synthesis during division.
Quiescence in Bacteria is Regulated by Peptidoglycan Cues
Uropathogenic E. coli is responsible for the majority of urinary tract infections. We investigate how this organism enters a QUIESCENT, non-proliferative and antibiotic-tolerant state, which may allow it to evade killing by antibiotics or the immune system during infection. Our work is focused on understanding how external cues, including peptidoglycan fragments, stimulate proliferation of E. coli from the quiescent state.
Amyloids, Aggregates and Protein Homeostasis
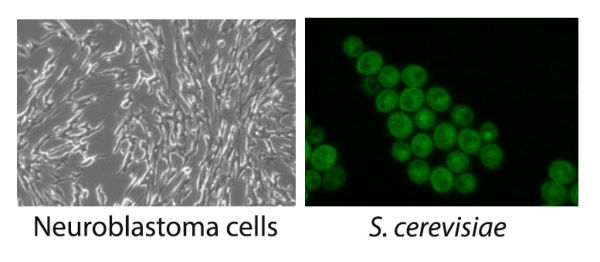
Protein homeostasis in differentiated neurons, yeast and bacteria
Chaperone-mediated remodeling and proteolysis
Molecular chaperone proteins maintain intracellular protein homeostasis in all living organisms. When these homeostatic mechanisms are disrupted, cells are less able to cope with environmental stress and maintain normal physiology and function. We study how proteins misfold and aggregate, and how molecular chaperones promote reactivation from aggregates and amyloids and partner with proteases for degradation in vivo and in vitro.
~~~~~
Protein homeostasis in bacteria, yeast & differentiated neurons:
Key cellular events under investigation in multiple organisms that are proteolytically regulated include cell division, toxin-antitoxin systems, dormancy, prion inheritance and clearance of amyloids and other aggregates.
ATP-dependent Protein Machines
Performing mechanical work at the angstrom scale
Reconstituting cell machinery
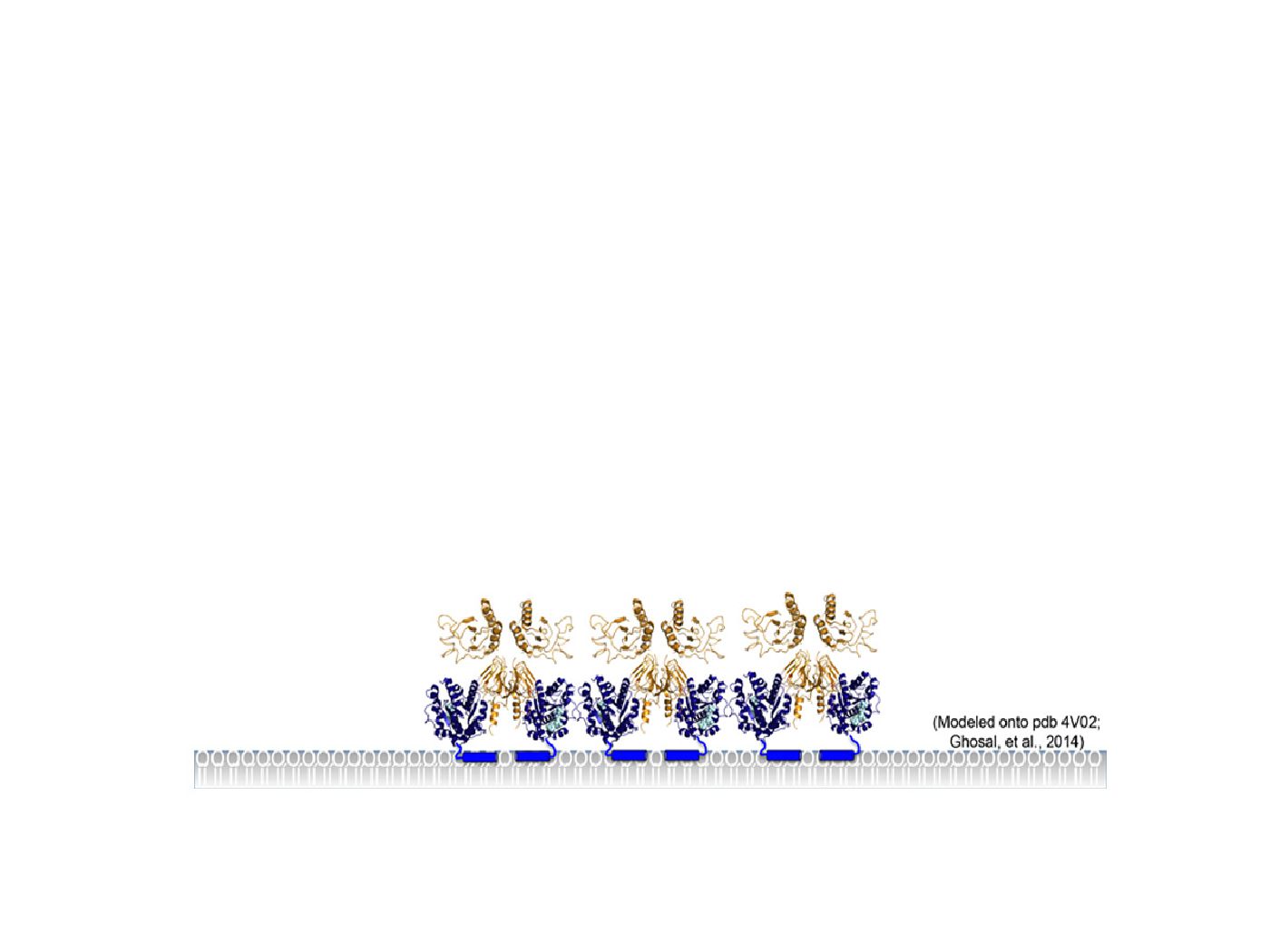
Molecular pathways are often highly complex with specific mechanisms being governed by multiple protein-protein interactions and protein-lipid interactions, and further regulated by enzymatic activities such as ATP or GTP hydrolysis. We take a reductionist approach to investigate each biochemical event in vitro. With an ever-expanding library of purified proteins and lipids we employ a combination of classical biochemical, biophysical and high resolution microscopic techniques to investigate specific molecular events. Recently, we have been investigating the polar oscillation of the Min system, direct interactions between FtsZ, FtsA and the lipid bilayer, and targeting and degradation of antitoxins, including MqsA, and cell division proteins by the AAA+ proteases ClpXP and Lon.


SHARE THIS PAGE!